About
What is eggplant?
eggplant is a computational method designed to enable the transfer
of observed spatial data such as for example - but not limited to - gene
expression to a CCF (common coordinate framework).
The method relies on spatial landmarks present in both the observed data as well as the reference we seek to transfer this to. In short, the spatial distribution of a FOI (feature of interest) is considered as a multivariate function of the distance to each landmark:
Where \(y_i\) is the FOI value at the i:th observation, and \(X_{ij}\) is the distance between the j:th landmark and the i:th observation. To learn this function (\(f\)) we use Gaussian Process Regression.
How does it work?
The method is more thoroughly described in it’s associated publication, but to briefly outline the steps, we will use the same flowchart as is presented in Figure 1A.
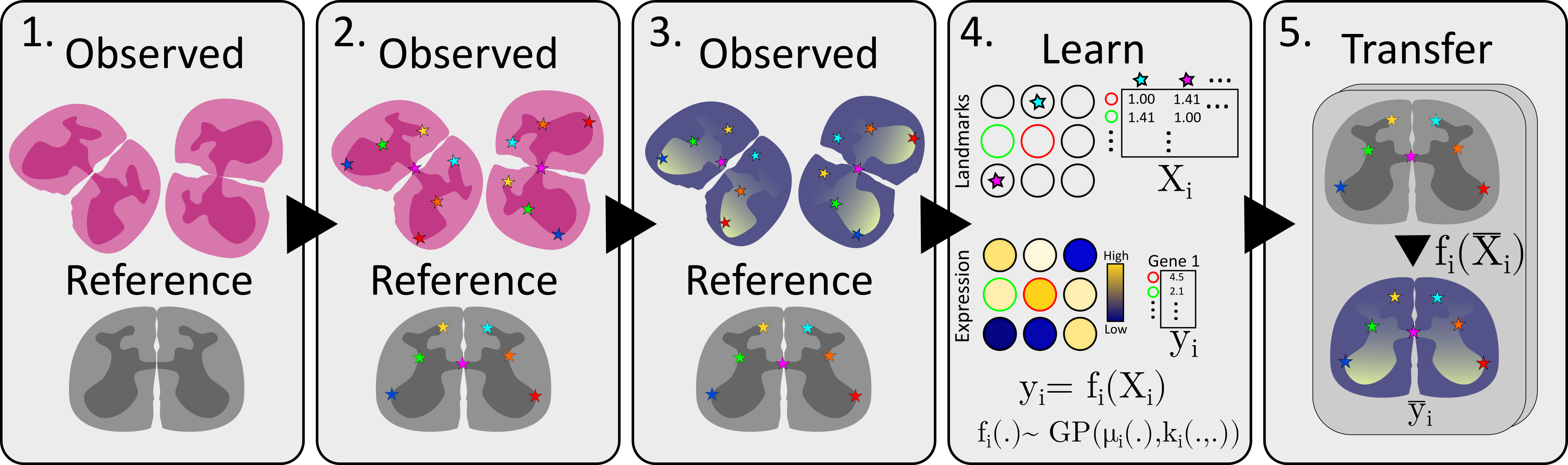
Construct/choose a reference and select samples to transfer information from
Chart the landmarks, i.e., annotate common landmarks
Select feature of interest, e.g., expression of your favorite gene
Learn a transfer function relating landmark distances to expression
Transfer information to the reference, apply transfer function to each location in the reference
To elaborate a bit more on the flow of information from observed data to the reference we refer to the image below.
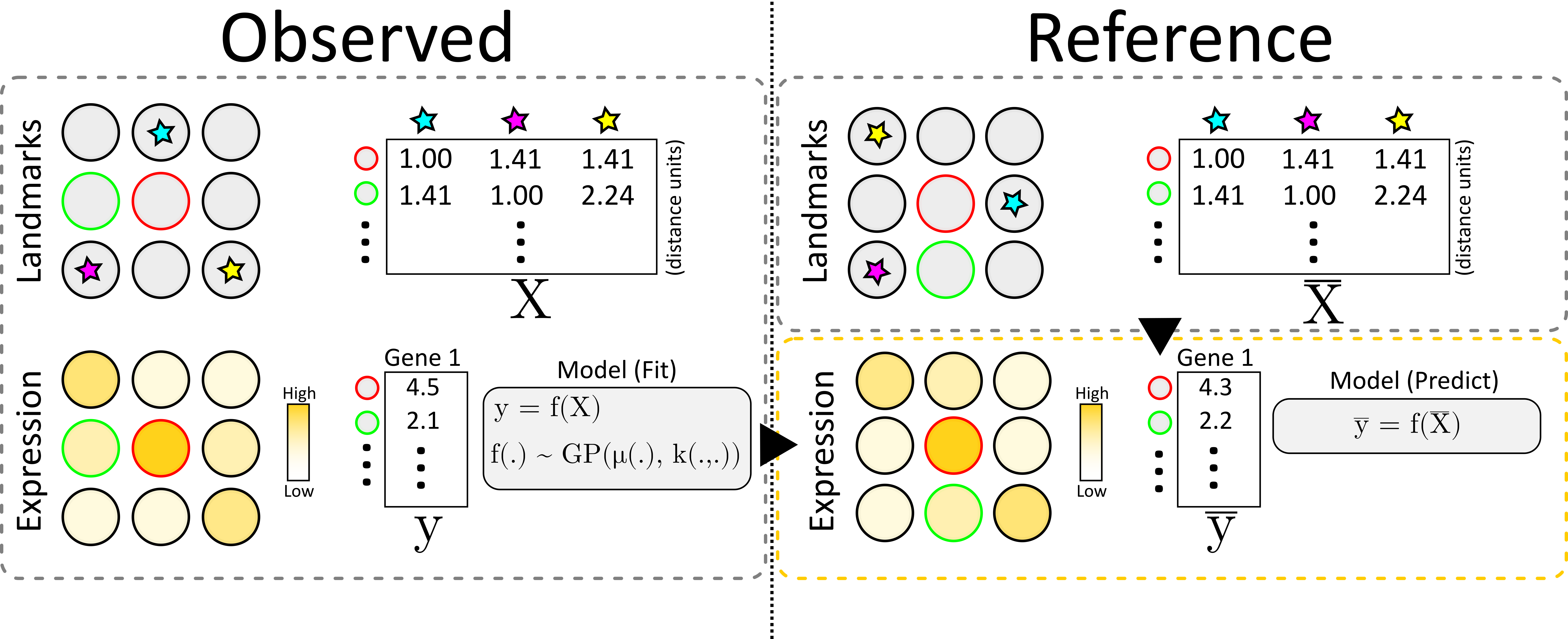
We start by creating a distance matrix \(X\) for the observed data, containing the distance from every observation to each landmark. Next, we also create the feature vector \(y\) containing the values for our feature of interest at each location. We model \(y\) as a function (\(f\)) of \(X\) assume that this function is distributed according to a Gaussian Process, and thus fit it accordingly.
To transfer the information, we first create a similar distance matrix to \(X\) but for all of the locations in our reference, this is denoted \(\bar{X}\). Finally, we apply the (learnt) function \(f\) to \(\bar{X}\), rendering a new feature vector \(\bar{y}\) which represents an estimate of the feature values at each of the reference locations.
Why the name eggplant?
Aside from being a delicious vegetable, the name is also an acronym for
effortless generic GP landmark transfer